If your sample contains proteins of interest that are less than 20 kDa, you can download a protocol that explains how to detect synthetic peptides or small molecular weight proteins using SDS-PAGE/Western blotting.
Here are some tips to help ensure your Western performs optimally.
If you want to validate your antibodies, here are some useful tips:
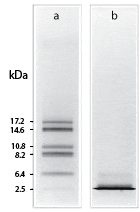
Shown is the resolution of cyanogen bromide fragments of myoglobin by (a) Tricine–SDS-PAGE and (b) Laemmli–SDS-PAGE using 10% T, 3% C gels.
Tricine-SDS-PAGE is a commonly used method for separating proteins in the mass range of 1-100 kDa. It is the preferred electrophoretic system for the resolution of proteins smaller than 30 kDa.
However, it can be difficult to detect small peptides using this method. In this case, a Tris-tricine gel may provide better resolution. If you only need to detect the peptide, Mass Spec is the best way to confirm its identity.
Smaller peptides bind less Coomassie brilliant blue than larger proteins, which makes them harder to detect by Coomassie staining or silver staining. If you want to see your peptide on the gel, you can try loading more samples. However, changing the gel percentage won't help much, unless you think your peptide migrated out of the gel. Instead, you can increase the percentage of the crosslinker in the regular 17% gel. Also, raising the pH of your resolving gel to 9.5 as compared to your regular 8.8, and adding urea (4-8M) can help sharpen bands.
If you plan to use a Western blot, which is a more sensitive detection method, it is better to use Western instead of gel staining. However, it can still be challenging to detect small peptides efficiently by the conventional Western blotting method because the peptides readily detach from the blotted membrane. To improve the detection, you can try using two pieces of membrane, a shorter time of transfer (less than 1 hour at 200 mA), and a 0.2um pore. Semi-dry transfer for 15-20 minutes at the recommended current density (mA/cm2) for the apparatus may also work for most of the small peptides.
Another method that can be used to shorten the time required for immunodetection steps is vacuum-assisted detection. Please check this reference: A new approach to detect small peptides clearly and sensitively by Western blotting using a vacuum-assisted detection method.
If you can plan ahead and synthesize a control small peptide labeled with biotin, you can monitor the transfer process and its ability to bind the membrane with streptavidin-conjugated HRP.
For more information on Tricine-SDS-PAGE, including efficient methods for Coomassie blue staining, silver staining, and electroblotting, please download this protocol.
Download the Protocol
Calculating peptide concentration is not the same as determining peptide purity. Purity is measured by HPLC and indicates the presence or absence of contaminants with undesired sequences. On the other hand, peptide content only gives information on the percentage of total peptide versus total non-peptide components independently of the presence of multiple peptides.
Net peptide content can be accurately determined by performing amino acid analysis or UV spectrophotometry. It is difficult to determine the actual peptide concentration based on the weight of the lyophilized peptide. Lyophilized peptides may contain 10-70% water and salts by weight, with more hydrophilic peptides generally containing more bound water and salts compared to hydrophobic peptides.
If the peptide contains tryptophan (W) or tyrosine (Y) residues, its concentration can be conveniently determined based on the extinction coefficient of these residues. The molar extinction coefficients of chromophoric residues at 280 nm at neutral pH using a 1-cm cell are: tryptophan 5560 AU/mmole/ml and tyrosine 1200 AU/mmole/ml. The overall molar extinction coefficient of the peptide depends on the types and number of these choromophoric residues in the sequence.
The following steps can be used for the calculations: