| 6213 |
| Peptide |
| Biotin-Ahx-GGSENLYFQSYVGG-{Nle}-P-{D-Phe}-R-{D-Trp}-FKAVGKKRR |
| |
| 4mg |
| >95% |
| |
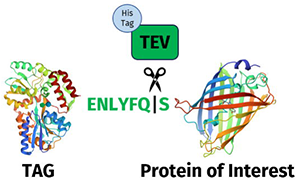
TEV Protease and Its Unique Sequence Specificity:
Tobacco Etch Virus (TEV) protease is a highly sequence-specific cysteine protease that cleaves precisely at the consensus sequence ENLYFQS, where ‘’ marks the peptide bond cleaved between glutamine (Q) and serine (S). This specificity has made TEV protease a powerful tool in biotechnology, particularly for processing recombinant proteins. Its precise cleavage pattern ensures that it removes affinity tags or unwanted sequences without additional non-specific cuts, preserving the integrity of target proteins and providing high reproducibility in various experimental conditions.
Due to its precision, TEV protease is invaluable in protein production, where tags are often used to facilitate purification but must be removed from the final product. TEV’s ENLYFQS recognition site enables researchers to cleave these tags accurately, yielding proteins with native sequences essential for structural and functional studies. Additionally, TEV protease plays a critical role in studying protein-protein interactions by tagging proteins with ENLYFQS sites in biosensors and conducting enzyme assays to investigate molecular mechanisms within cellular environments.
TEV protease’s structural design, particularly its catalytic triad (His46, Asp81, Cys151), enables high selectivity and has inspired extensive research on improving its function. Through techniques like molecular dynamics (MD) simulations, scientists have created modified TEV variants that increase efficiency and reduce self-cleavage tendencies. These studies show how structural adaptations around the active site enhance catalytic function, broadening TEV's use in more demanding biotechnological applications and guiding efforts in enzyme engineering.
The ENLYFQS sequence’s uniqueness and the ongoing innovations in TEV protease design continue to expand its utility across both basic research and therapeutic development, establishing it as a reliable and versatile tool for precise protein processing in laboratory settings.
 https://www.lifetein.com
100 Randolph Road, Suite 2D,
Somerset
USA
New Jersey
08873
https://www.lifetein.com
100 Randolph Road, Suite 2D,
Somerset
USA
New Jersey
08873
 https://www.lifetein.com
100 Randolph Road, Suite 2D,
Somerset
USA
New Jersey
08873
https://www.lifetein.com
100 Randolph Road, Suite 2D,
Somerset
USA
New Jersey
08873