Amino Acid Properties and Structure
Amino acids are molecules that contain an amine group, a carboxylic acid group, and a side-chain that varies among amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen. R stands for the 20 possible different side chains. The R group might be as simple as a hydrogen atom, or as complex as a carbon chain with functional groups. The physical and chemical properties of the R side chains determine the unique characteristics and functions of each particular amino acid, and thus the entire protein.
Peptide Software and Property Calculators:
1. Protein structure prediction software: RoseTTAFold is an advanced protein structure prediction software developed by the Institute for Protein Design. Powered by multiple neural networks, it rapidly and accurately predicts 3D protein structures directly from amino acid sequences—reducing a process that once took years in the lab to just minutes. Building on this capability, RoseTTAFold All-Atom models complex biological systems at atomic resolution, enabling detailed predictions of protein–DNA interactions, drug–receptor binding, and more. This breakthrough tool is accelerating discoveries across biology, medicine, and biotechnology.
2. PROTEIN CALCULATOR v3.4 from Scripps
3. Proteomics from ExPASy
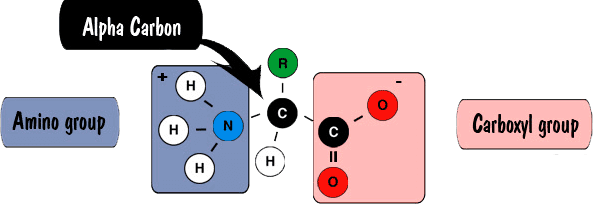
The Function of Amino Acids in a Protein Structure
- Alanine is not particularly hydrophobic, and is non-polar. However, because it contains a normal C-beta carbon the conformations its backbone can adopt are limited. Generally, Ala plays a non-critical role in proteins. Its side chain is very unreactive; therefore, it is rarely involved in protein function directly. However, it can play a role during substrate recognition or specificity, and in particular in the interactions of Ala with non-reactive atoms such as carbon.
Name
Abbreviation |
M.W. |
pI |
CAS Registry Number |
Structure / formula |
3D Structure |
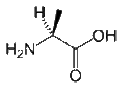
| Alanine |
89.09 |
6 |
56-41-7 |
 |
 |
| Ala A |
CH3-CH(NH2)-COOH |
|
- Arginine is a positively charged, polar amino acid. These amino acids reside preferentially on the outside of proteins.Arg commonly plays an important structural role. The part of its side chain closest to the peptide backbone is long and hydrophobic and contains carbon, whereas there is a positive charge at the end of the side chain. Therefore, Arg is commonly located where part of the side-chain is buried so that only the charged portion is oriented outside the protein. Arginines also play roles in salt-bridges, in which they pair with negatively charged amino acids (such as aspartate) to form stabilizing hydrogen bonds that are important for protein stability.
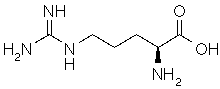
| Arginine |
174.2 |
11.15 |
74-79-3 |
 |
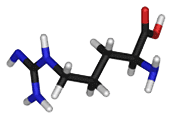 |
| Arg R |
HN=C(NH2)-NH-(CH2)3-CH(NH2)-COOH |
|
- Asparagine is a polar amino acid. Aspartate differs from asparagine only in that it contains an oxygen atom where there is an amino group in asparagine. Asn is generally located on the surface of proteins where it is exposed to an aqueous environment. It commonly plays roles in the active or binding sites of proteins. The polar side-chain can form interactions with other polar or charged atoms.
| Asparagine |
132.12 |
5.41 |
5794-13-8 |
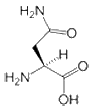 |
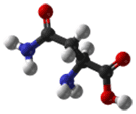 |
| Asn N |
H2N-CO-CH2-CH(NH2)-COOH |
|
- Aspartate (or Aspartic acid) is a polar, negatively charged amino acid. When they are buried within a protein, aspartates (and glutamates) commonly form salt-bridges by pairing with positively charged amino acids (such as arginine) to create hydrogen bonds that stabilize the structure of a protein. Asp is frequently involved in the active or binding sites of proteins. Its negative charge can interact with positively charged non-protein atoms, for example cations such as zinc. Asp has a shorter side-chain than the similar glutamate; therefore, it is slightly less flexible within a protein structure. The best example of Asp in active sites is in serine proteases such as trypsin, where it functions as part of a classical Asp-His-Ser catalytic triad.
| Aspartic acid |
133.1 |
2.77 |
56-84-8 |
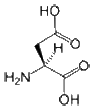 |
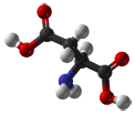 |
| Asp D |
HOOC-CH2-CH(NH2)-COOH |
|
- Cysteine: The role of cysteine in the structure of a protein depends on the cellular location of the protein. When found in extracellular proteins, Cys residues frequently form disulfide bonds, in which pairs of cysteines are oxidized to form covalent bonds. Disulfide bonds generally stabilize the structure of a protein. In fact, the structure of several extracellular proteins is determined almost entirely by the topology of multiple disulfide bonds. In contrast, the reducing environment found intracellularly makes disulfide bond formation very unlikely. Disulfide bonds are also rare within membranes, although membrane proteins can contain disulfide bonds in their extracellular domains. Nevertheless, Cys can still play an important structural role in an intracellular environment. Its sulfhydryl side-chain can bind readily to metals such as zinc; therefore, Cys (and other amino acids such as His) are very common in metal-binding motifs such as zinc fingers. Metal binding can also be important for the function of certain enzymes, such as metal proteases. Cys is also very common in the active and binding sites of proteins. It can also function as a nucleophile (i.e. the reactive center of an enzyme); probably the best-known example of this is in cysteine proteases such as caspases or papains, where Cys is the key catalytic residue, albeit facilitated by His and Asn.
| Cysteine |
121.15 |
5.02 |
52-90-4 |
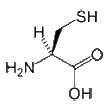 |
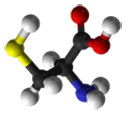 |
| Cys C |
HS-CH2-CH(NH2)-COOH |
|
- Glutamine is a polar amino acid. Generally, Gln prefers to be on the protein surface where it is exposed to aqueous environments. It is commonly involved in the active or binding sites of proteins. In addition, its polar side-chain interacts with other polar or charged atoms.
| Glutamine |
146.15 |
5.65 |
56-85-9 |
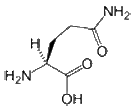 |
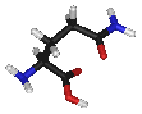 |
| Gln Q |
H2N-CO-(CH2)2-CH(NH2)-COOH |
|
- Glutamate (or Glutamic acid) is a negatively charged, polar amino acid. Generally, Glu prefers to be localized to the protein surface, where it is exposed to an aqueous environment, because it is both charged and polar. When buried within a protein it commonly forms salt-bridges by pairing with a positively charged amino acid to create hydrogen bonds that stabilize the structure of a protein. Glu is commonly involved in the active or binding sites of proteins. Its negative charge means that it can interact with positively charged non-protein atoms, including cations such as zinc. It can also perform a similar role to aspartate in the catalytic site of proteases or lipases under certain conditions.
| Glutamic acid |
147.13 |
3.22 |
56-86-0 |
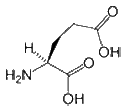 |
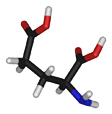 |
| Glu E |
HOOC-(CH2)2-CH(NH2)-COOH |
|
- Glycine is a unique amino acid because it contains a hydrogen atom instead of a carbon as its side chain; this results in a high level of conformational flexibility. As such, Gly can be located in parts of a protein structure that are too small for all other amino acids. Glycine can bind to phosphate groups because of its hydrogen side chain. Changing a conserved glycine residue to any other amino acid has a significant effect on the stability and structure of a protein.
| Glycine |
75.07 |
5.97 |
56-40-6 |
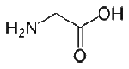 |
 |
| Gly G |
NH2-CH2-COOH |
|
- Histidine is a polar amino acid that has a pKa close to that of physiological pH. As such, the removal and addition of protons from its side chain is relatively straightforward, which can alter its side chain from neutral to positively charged. Because of its flexibility, it is unclear whether His prefers to be buried in the protein core or exposed to an aqueous environment. Therefore, it is an ideal residue for the center of a functional protein. His is the most common amino acid that is located in the active or binding site of a protein. It is very common in metal binding sites (e.g. zinc), where it commonly functions with cysteines or other amino acids.
| Histidine |
155.16 |
7.47 |
71-00-1 |
 |
 |
| His H |
N=C-NH-C=C-CH2-CH(NH2)-COOH |
|
- Isoleucine is an aliphatic and hydrophobic amino acid; therefore, it prefers to be buried within the hydrophobic core of proteins. Like valine and threonine, isoleucine is C-beta branched. Although most amino acids contain only one non-hydrogen atom attached to their C-beta carbon, these amino acids all contain two. Therefore, the number of conformations that their main chains can adopt is limited. For example, it is more difficult for Ile to adopt an alpha-helical conformation, whereas it is simple (and perhaps preferred) for them to reside in beta-sheets. The side chain of Ile is very non-reactive. Although it plays a direct role in protein function only rarely, it can play be involved in substrate recognition. Generally, hydrophobic amino acids can be important for the recognition and binding of hydrophobic ligands such as lipids.
| Isoleucine |
131.17 |
5.94 |
73-32-5 |
 |
 |
| Ile I |
CH3-CH2-CH(CH3)-CH(NH2)-COOH |
|
- Leucine is an aliphatic and hydrophobic amino acid that prefers to be buried within the hydrophobic core of a protein. It is more commonly found in alpha-helices than in beta-strands. Because it has a very non-reactive side chain it is rarely involved in protein function directly. Nevertheless, it can play a role in substrate recognition, for example the recognition of and binding to hydrophobic ligands such as lipids.
| Leucine |
131.17 |
5.98 |
61-90-5 |
 |
 |
| Leu L |
(CH3)2-CH-CH2-CH(NH2)-COOH |
|
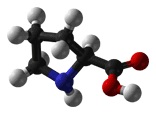
- Lysine is a positively charged and polar amino acid that commonly plays a very important structural role. The part of the side chain nearest to the peptide backbone is long and hydrophobic and contains carbons, whereas the end of its side chain is positively charged. Lys is commonly located so that much of its side chain is buried, and only the charged portion is oriented toward the outside of the protein. Lys commonly plays a role is salt bridge formation, where it pairs with a negatively charged amino acid (such as aspartate) to create the hydrogen bonds that stabilize protein structure. It is also found frequently in the active or binding sites of proteins. The positively charged amino group on the side chain of Lys can also form hydrogen bonds with negatively charged non-protein atoms (e.g. anions or carboxylate groups).
| Lysine |
146.19 |
9.59 |
39665-12-8 |
 |
 |
| Lys K |
H2N-(CH2)4-CH(NH2)-COOH |
|
- Methionine is a hydrophobic amino acid, and so it is commonly buried in the hydrophobic core of a protein. Because its side chain is fairly non-reactive it is rarely involved in protein function directly. Nevertheless, it can play a role in the recognition of and binding to hydrophobic ligands such as lipids, similar to other hydrophobic amino acids. However, Met also contains a sulfur atom that can bind to atoms including metals. However, although the sulfur atom in Cys is connected to a hydrogen atom and is thus reactive, the sulfur in Met is connected to a methyl group, which limits its role in protein function.
| Methionine |
149.21 |
5.74 |
63-68-3 |
 |
 |
| Met M |
CH3-S-(CH2)2-CH(NH2)-COOH |
|
- Phenylalanine is an aromatic and hydrophobic amino acid that prefers to be buried within the hydrophobic core of proteins. The side chain of Phe is fairly non-reactive; therefore, it is involved only rarely in protein function directly, although it can play a role in recognizing hydrophobic ligands and substrates such as lipids. Aromatic amino acids such as Phe can also interact with non-protein ligands that also contain aromatic groups via stacking interactions, and can also bind to polyproline-containing peptides, for example in SH3 or WW domains.
| Phenylalanine |
165.19 |
5.48 |
63-91-2 |
 |
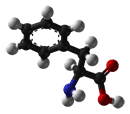 |
| Phe F |
CH3-CH2-CH(NH2)-COOH |
|
- Proline is the only amino acid whose side chain is connected to the protein backbone twice, which forms a five-membered nitrogen-containing ring. Therefore, proline is an imino acid because it contains an NH2+ group instead of the more common NH3+ group. As such, Pro is unable to adopt some of the main chain conformations that are formed easily by all other amino acids. Therefore, it could be considered to be the opposite of glycine, which can adopt several main chain conformations. Proline is often located at very sharp turns in protein structures in locations where a polypeptide chain changes direction. It can also introduce turns and kinks into alpha-helices since it cannot adopt a normal helical conformation. Pro is commonly found on the surface of a protein, where it plays an important role in molecular recognition, particularly during intracellular signaling. Specifically, protein domains such as WW and SH3 bind to specific proline-containing peptides that form key parts of multiple important signaling cascades. Because its side chain is very non-reactive and it has difficulty adopting many main chain conformations, Pro is very rarely involved in the active or binding sites of proteins.
| Proline |
115.13 |
6.3 |
147-85-3 |
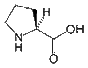 |
 |
| Pro P |
NH-(CH2)3-CH-COOH |
|
- Serine is a slightly polar amino acid that can be located either within the interior or on the surface of a protein. Its small size means that it is found commonly in tight turns on the protein surface, which allows the hydroxyl oxygen of the Ser side chain to form a hydrogen bond with the protein backbone, effectively mimicking proline. Ser is also relatively common in the functional centers of proteins. Its hydroxyl group is fairly reactive, and can form hydrogen bonds with a variety of polar substrates. An important and common role for Ser (as well as Thr and Tyr) within intracellular proteins is phosphorylation. Phosphate groups are commonly attached to Ser to facilitate signal transduction pathways. Although this role of serine could often be replaced by threonine, it is unlikely to be replaced by tyrosine, because the protein kinases that catalyze the phosphorylation reactions are highly specific. Specifically, tyrosine kinases generally do not phosphorylate serines and threonines, and vice-versa.
| Serine |
105.09 |
5.68 |
56-45-1 |
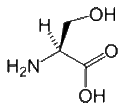 |
 |
| Ser S |
HO-CH2-CH(NH2)-COOH |
|
- Threonine can reside either within a protein interior or on its surface. Thr is restricted in terms of the number of conformations that its main chain can adopt. Although it is difficult for it to form an alpha-helical conformation, it is easy (and perhaps preferred) for it to form beta-sheets. Thr is found commonly in the functional centers of proteins. Its hydroxyl group is fairly reactive, and can form hydrogen bonds with a variety of polar substrates. A common role for Thr (as well as Ser and Tyr) in intracellular proteins is to be phosphorylated, whereby protein kinases attach phosphate groups to Thr during signal transduction pathways. Although threonine could be replaced by serine, it is unlikely to be replaced by tyrosine because the enzymes that catalyze phosphorylation reactions are highly specific.
| Threonine |
119.12 |
5.64 |
72-19-5 |
 |
 |
| Thr T |
Ph-CH(OH)-CH(NH2)-COOH |
|
- Tryptophan is an aromatic and hydrophobic amino acid that is preferentially buried within the core of proteins. Tryptophan can form stacking interactions with the aromatic side-chains of amino acids. Because it contains nitrogen rather than carbon in its aromatic ring, Trp is more reactive than Phe, but less reactive than Tyr. Similar to other aromatic amino acids, it can form stacking interactions with non-protein ligands that themselves contain aromatic groups. Trp and other aromatic amino acids can also play a role in binding to polyproline-containing peptides, for example those found in SH3 or WW domains.
| Tryptophan |
204.23 |
5.89 |
73-22-3 |
 |
 |
| Trp W |
Ph-NH-CH-C-CH2-CH(NH2)-COOH |
|
- Tyrosine is an aromatic amino acid that is partially hydrophobic and is preferentially buried within a protein hydrophobic core. Tyrosine forms stacking interactions with other aromatic side-chains. Because it contains a reactive hydroxyl group it is far more likely than Phe to interact with non-protein atoms. Similar to other aromatic amino acids, Tyr is involved in binding to polyproline containing-peptides, such as in SH3 or WW domains. A common role for tyrosine (as well as serine and threonine) is phosphorylation. During intracellular signaling, protein kinases commonly attach phosphate groups to Tyr to facilitate the signal transduction process.
| Tyrosine |
181.19 |
5.66 |
60-18-4 |
 |
 |
| Tyr Y |
HO-p-Ph-CH2-CH(NH2)-COOH |
|
- Valine is an aliphatic and hydrophobic amino acid that is commonly buried within the hydrophobic cores of proteins. Its side chain is very non-reactive, and so it is directly involved in protein function only rarely. Nevertheless, it can play a role in recognizing substrates, as well as recognizing and binding to hydrophobic ligands such as lipids.
| Valine |
117.15 |
5.96 |
72-18-4 |
 |
 |
| Val V |
CH3-CH(CH2)-CH(NH2)-COOH |
|
Amino Acid Classification
Amino acids can be classified according to:
- Number of amino and carboxyl groups in the amino acid
- Aliphatic amino acids: Alanine, Valine, Leucine, Isoleucine, Proline. Aliphatic implies that the protein side chain contains only carbon or hydrogen atoms.
- Aromatic amino acids: Phenylalanine, Histidine, Tyrosine and Tryptophan. A side chain is aromatic when it contains an aromatic ring system. Generally, aromatic ring systems are planar, and electons are shared over the whole ring structure.
- Hydroxy amino acids: Serine, Threonine, Tyrosine.
- Sulphur-containing amino acids: Methionine and Cysteine.
- Heterocyclic amino acids as histidine, tryptophan.
- Imino acids as proline and hydroxyproline.
- Nutritional: Essential amino acids: Phenylalanine, Treptophan,Histidine, Methionine, Threonine, Leucine, Valine, Isoleucine, Arginine, and Lysine. Non essential amino acids: Glycine, alanine, serine, tyrosine, proline, hydroxyl proline, cystiene, aspartic acid and glutamic acid
- Metabolic: glucogenic or ketogenicamino acids, and charge and polarity of side chain.
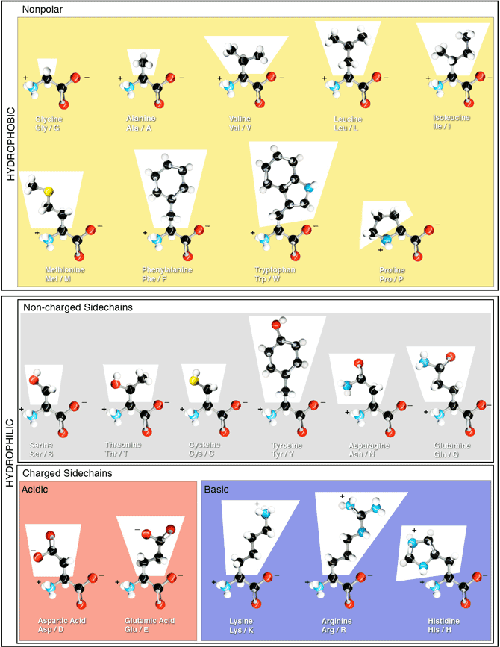
Amino Acid Properties
| Amino Acid |
Code |
Hydropathy |
Charge |
pKa, NH2 |
pKa, COOH |
pK(R) |
Solubility |
| Arginine |
R |
hydrophilic |
+ |
9.09 |
2.18 |
13.2 |
71.8 |
| Asparagine |
N |
hydrophilic |
N |
8.8 |
2.02 |
|
2.4 |
| Aspartate |
D |
hydrophilic |
- |
9.6 |
1.88 |
3.65 |
0.42 |
| Glutamate |
E |
hydrophilic |
- |
9.67 |
2.19 |
4.25 |
0.72 |
| Glutamine |
Q |
hydrophilic |
N |
9.13 |
2.17 |
|
2.6 |
| Lysine |
K |
hydrophilic |
+ |
10.28 |
8.9 |
2.2 |
|
| Serine |
S |
hydrophilic |
N |
9.15 |
2.21 |
|
36.2 |
| Threonine |
T |
hydrophilic |
N |
9.12 |
2.15 |
|
freely |
| Cysteine |
C |
moderate |
N |
10.78 |
1.71 |
8.33 |
freely |
| Histidine |
H |
moderate |
+ |
8.97 |
1.78 |
6 |
4.19 |
| Methionine |
M |
moderate |
N |
9.21 |
2.28 |
|
5.14 |
| Alanine |
A |
hydrophobic |
N |
9.87 |
2.35 |
|
15.8 |
| Valine |
V |
hydrophobic |
N |
9.72 |
2.29 |
|
5.6 |
| Glycine |
G |
hydrophobic |
N |
9.6 |
2.34 |
|
22.5 |
| Isoleucine |
I |
hydrophobic |
N |
9.76 |
2.32 |
|
3.36 |
| Leucine |
L |
hydrophobic |
N |
9.6 |
2.36 |
|
2.37 |
| Phenylalanine |
F |
hydrophobic |
N |
9.24 |
2.58 |
|
2.7 |
| Proline |
P |
hydrophobic |
N |
10.6 |
1.99 |
|
1.54 |
| Tryptophan |
W |
hydrophobic |
N |
9.39 |
2.38 |
|
1.06 |
| Tyrosine |
Y |
hydrophobic |
N |
9.11 |
2.2 |
10.1 |
0.038 |
Fluorophore list including absorption and emission peak values
|
Fluorophore |
Absorption |
Emission |
| Fluorescein |
495 nm | 517 nm |
| FITC |
495 nm | 517 nm |
| Rhodamine 110 |
497 nm | 520 nm |
| Rhodamine B |
543 nm | 565 nm |
| 5-TAMRA-MeOH |
543 nm | 567 nm |









































