Please click here to receive a peptide quote now!
Did you know that a peptide length of eight residues is adequate to identify elements of structure in PDB studies? Send us your peptide sequence for bioinformatics analysis and order your 8mer Now!
Peptides often have conformational preferences. Many short peptides in solution can adapt their native secondary structures: α helices and β hairpins. To date, such studies are mainly focused on selected peptides that are expected to fold. Many models assume that peptide fragments with preferred conformations are responsible for nucleating the folding process. In this study (DOI: 10.1371/journal.pcbi.0020027), the researchers systematically generate a series of 8-mer peptide fragments with overlapping sequences from the original protein sequence. The 8-mers were chosen because this length is adequate to identify elements of structure in PDB studies. In this study, the researchers got the peptide fragments from a series of well-characterized proteins: protein G, protein L, protein A, and chymotrypsin inhibitor.
Do peptides have native-like conformations? Of the 133 peptides studied, the researchers found that 48 peptides have the structural bias as structured peptides. In the protein G fragments, they find 8 structured peptides have adopted a stable helical-turn conformation. Out of the 48 structured peptides, only five differ significantly from the native structure. So 41 peptides fold into approximately their native conformations. Why are there only a small number of the peptides structured? By using very short peptides, most of the hydrophobic clustering, or helical hydrogen bonds may have been eliminated. So, it is important to choose the specific sequence motifs with a high-confidence prediction. A well-designed structure-prediction system with a pragmatic mix of bioinformatics and physical modeling is the key to the peptide design.
Order your 8-mer now! We will analyze the peptide structure and suggest the sequences for you.
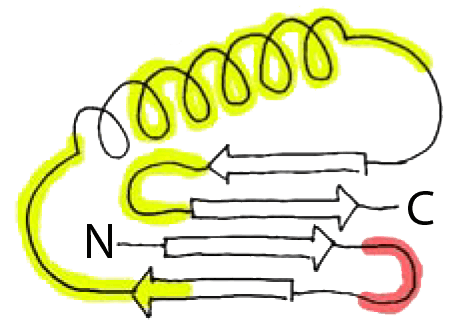
Distribution of Kinks in the Protein G Structure: coils (α helices); notch (hydrogen-bonded β-turns); arrows (β strands); Yellow (helical-turn); red (reverse-turns).
Did you know that a peptide of 35 residues is the smallest folded domain of a naturally occurring protein? How does the amino acid sequence of a protein chain determine and remain its 3D folded state? How do small proteins fold?
In a recent study, Rocklin et al. designed sequences intended to fold into desired structures. The novel proteins may be useful in bioengineering or pharmacological applications. Check the paper from here: https://goo.gl/Tregb7
Many small proteins or miniproteins are peptides shorter than 40-50 residues with stable folding that contain secondary structure elements such as α helices and β strands.
An autonomously folding, 35 residues, thermostable subdomain (HP36) of the villin headpiece, is the smallest folded domain of a naturally occurring protein. So, polypeptides simplify the protein-folding problem. It allows in-depth examinations of sequence-structure-stability relationships without using the complex larger proteins.
In this study, Rocklin et al. identified over 2,500 new stable designed proteins. The study systematically examined how sequence determines folding and stability. Many proteins are stable without disulfides or metal coordination and are more stable than any comparably-sized monomeric proteins found in the PDB. Many stable designs have the structure of ααα or βαββ. Stable designs buried more nonpolar surface area than did unstable designs. It was found that amino acids (Asp, Ser, Thr, and Asn) are favorable for capping helices. But these amino acids (Asp, Ser, Thr, and Gly) are unfavorable within helices.
In conclusion, to get a functional peptide that potentially folds naturally, a short peptide of at least 8mer or a long peptide of 35-45mer with the predicted α helices and β strands may be necessary. The stabilizing roles for aromatic residues at surface-exposed sites may be important because of the noncovalent interactions.
