There are many ways to design improved peptides: site-directed mutagenesis, computational design approaches, synthetic libraries, template-assisted methodologies, and mechanism-based strategies.
- Site-directed mutagenesis:
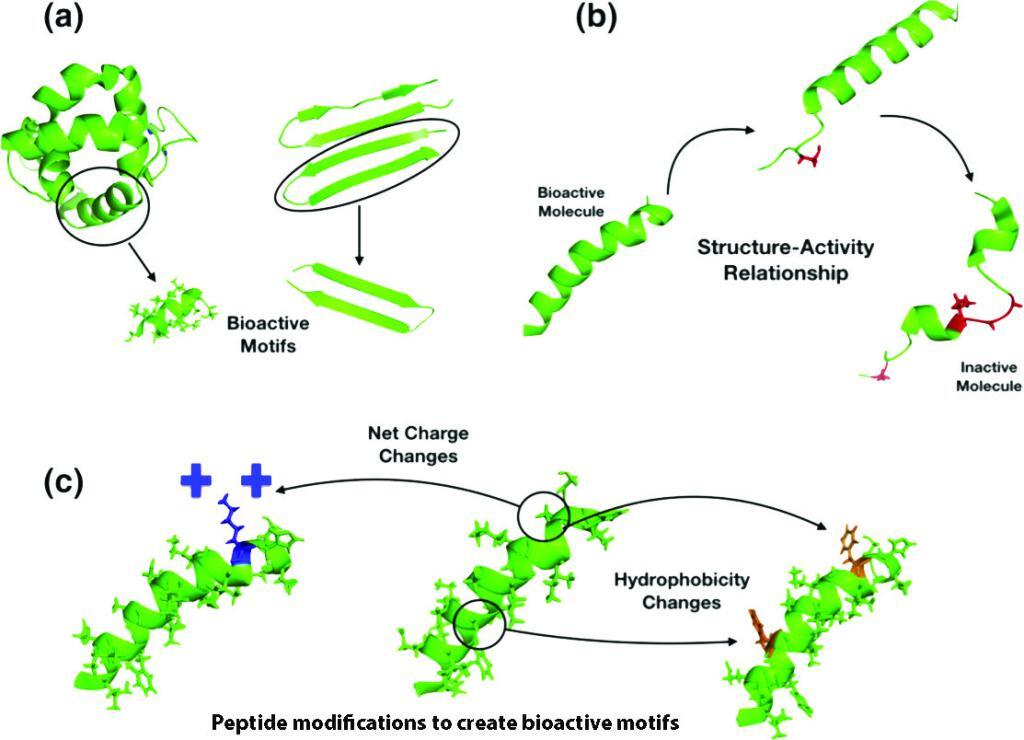
Adding, deleting, or replacing one or more amino acid residues are useful approaches to re-engineer a peptide. The alanine or lysine scanning are examples of valuable strategies, as they allow for coverage of all positions within a peptide chain, thus making it possible to analyze the effect of all amino acid side chains on structure and function. - De novo design:
Peptides present both basic and hydrophobic residues in a given sequence. The antimicrobial peptides favor an amphipathic structure. The helix-stabilizing residues such as leucine, alanine, valine, isoleucine, lysine, and arginine, or prototypical destabilizing residues such as proline and glycine are frequently introduced to manipulate the peptide structure. The design can also base on the native bioactive peptides as the template-assisted approach. - Synthetic libraries:
A synthetic combinatorial library is practical to scan for the active binding site. The positional scanning or iterative approaches are costly and time-consuming. - Computational methods:
The antimicrobial databases are widely available. The design can be based on the bioinformatics tools, statistical modeling, SAR studies, genetic algorithms or deep learning. The sequence motifs, structure, net charge, hydrophobicity, amphipathicity, and unnatural modifications are essential guidelines for the peptide design.
