An old adage says: “Show me your friends, and I'll know who you are.”
In the same way, identifying the compounds capable of interacting with a specific protein can reveal its function. Synthetic peptides are one of the approaches for detecting protein interactions.
An hsp70 (heat-shock protein of relative molecular mass 70K) can distinguish only unfolded forms of protein. To study the amino acid preferences, Gregory C. Flynn et. al. used the random-sequence peptides to fill the binding site of Binding immunoglobulin protein (BiP). It was found that the binding site of BiP shows considerable specificity.
Reference: Gregory C. Flynn, Jan Pohl, Mark T. Flocco & James E. Rothman. Peptide-binding specificity of the molecular chaperone BiP. 24 October 1991, Nature 353, 726-730 doi:10.1038/353726a0.
The simplest method for identifying the binding partners of a peptide is to use it as bait in an affinity pull-down experiment, and then detect its binding proteins directly. Pull-down assays are useful both for confirming the protein-protein interactions that were predicted using other techniques (e.g., co-immunoprecipitation) and also as initial screening tools to identify novel protein-protein interactions. Synthetic peptides are commonly used to verify suspected protein-protein interactions by disrupting the binding competitively.
Biotinylated peptides, which contain a specific functional domain, and their corresponding control native peptides are immobilized to avidin-conjugated resins. Samples are incubated with the resins. Then wash the resins to remove any unbound proteins. The bound proteins are eluted and analyzed using SDS-PAGE. The amount of modified versus unmodified bound peptide can then be compared to identify specific functions.
Biotinylated peptides can be synthesized chemically with specific modifications at a purity >80%. Ideally, the peptides should be ~15–20 amino acids; the modification should be located in the center of the sequence, and there should be a minimum of 6–8 flanking residues either side. Although biotin could be added to either the N- or C-terminus, we generally recommend the use of an N-terminal modification because it has a higher success rate, shorter turnaround time, and is easier to perform. In contrast, C-terminal modifications require the addition of an additional lysine residue.
Protocol used to Pull-Down Biotin-labeled Peptides
First, the resin to be used for peptide pull-down is generated by conjugating biotinylated peptides to avidin beads. Next, immobilized streptavidin beads are loaded with biotinylated peptide, and are then incubated with the cell lysates obtained from cells that were lysed in 1% (v/v) Nonidet P-40, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5 with protease and phosphatase inhibitors. Specifically, equal amounts of protein are incubated with the specific immobilized peptide of interest at 4°C for 6 h. The bound proteins are then eluted from the immobilized peptides by boiling in SDS sample buffer, or by cleavage using 50 mM dithiothreitol. Further analyses are then performed using eluates from pull-downs performed with the active and control bait peptides.
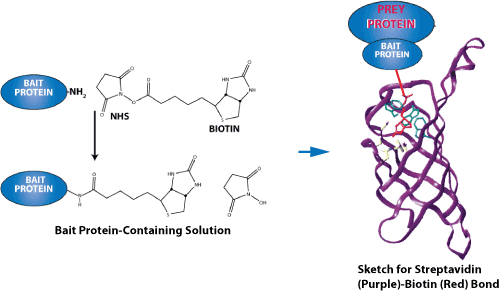
An image demonstrating the force of the streptavidin-biotin bond is shown below. Because they have an exceptionally high binding affinity, streptavidin-biotin and avidin-biotin pairs are two of the most prominent ligand-receptor pairs. Both proteins have a tetrameric structure; therefore, they can bind up to four ligands
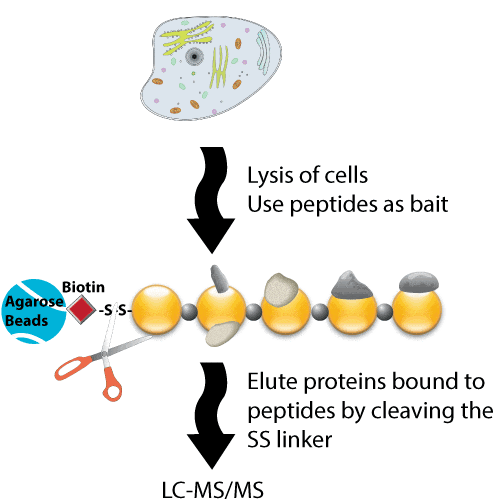
Peptides that correspond to specific regions that might be involved in protein-protein interactions can be synthesized. Proteins within the biological samples being analyzed then bind to the bait peptides, and are eluted, mixed, and analyzed. The eluted proteins are then digested using trypsin, and detected using mass spectrometry or SDS-PAGE. This technique is particularly useful for labeled cells. The reference described below provides an example of a method that used 13C-labeled cells. First, phosphorylated and non-phosphorylated peptides are synthesized separately. The 13C-labeled proteins were incubated with two related forms. The data allow a larger peak intensity corresponding to the C13-labeled form to be identified from the phosphorylated bait peptide. Therefore, this novel technique uses synthetic unmodified and modified peptides to allow a proteomic screen to be performed to identify direct protein binding partners. Importantly, this screening method is unbiased, highly specific, and reproducible, and requires only the presence of the binding partner in the cell lysates. The resulting data are also selective because only a limited number of proteins are retrieved from the large number of proteins contained within whole cell lysates.
Reference: A Novel Proteomic Screen for Peptide-Protein Interactions, J. Biol. Chem. 2004, 279:10756-10764, doi: 10.1074/jbc.M309909200
Public prtein-protein interaction databases
After the initial screen for protein–protein interactions a list of potential targets has been generated. The next step is to search the literature to find out if there are any proteins in the list that has been detected by other investigators as binding partners to the bait protein. The most accurate hot spot predictions are obtained for mutations involving Lysine. Lysine is a positively charged, polar amino acid. The side chain of lysine contains four carbon atoms which confer to it a partial hydrophobic character. Lysine is among the more frequent amino acids in hot spots.DIP (http://dip.doe-mbi.ucla.edu/),
IntAct (http://www.ebi.ac.uk/intact),
LifeTein is pleased to offer a free, comprehensive web-based peptide analysis tool. This peptide analysis tool will allow your research team to overcome common difficulties inherent in protein analysis and peptide antigen design. Try it now!
